\n",
"\n",
"\n",
"\n",
"\n",
"## The `hops` python module\n",
"\n",
"`hops` is a helper library for Hops that facilitates development by hiding the complexity of running applications and iteracting with services.\n",
"\n",
"Have a feature request or encountered an issue? Please let us know on github.\n",
"\n",
"### Using the `experiment` module\n",
"\n",
"To be able to run your Machine Learning code in Hopsworks, the code for the whole program needs to be provided and put inside a wrapper function. Everything, from importing libraries to reading data and defining the model and running the program needs to be put inside a wrapper function.\n",
"\n",
"The `experiment` module provides an api to Python programs such as TensorFlow, Keras and PyTorch on a Hopsworks on any number of machines and GPUs.\n",
"\n",
"An Experiment could be a single Python program, which we refer to as an **Experiment**. \n",
"\n",
"Grid search or genetic hyperparameter optimization such as differential evolution which runs several Experiments in parallel, which we refer to as **Parallel Experiment**. \n",
"\n",
"ParameterServerStrategy, CollectiveAllReduceStrategy and MultiworkerMirroredStrategy making multi-machine/multi-gpu training as simple as invoking a function for orchestration. This mode is referred to as **Distributed Training**.\n",
"\n",
"### Using the `tensorboard` module\n",
"The `tensorboard` module allow us to get the log directory for summaries and checkpoints to be written to the TensorBoard we will see in a bit. The only function that we currently need to call is `tensorboard.logdir()`, which returns the path to the TensorBoard log directory. Furthermore, the content of this directory will be put in as a Dataset in your project's Experiments folder.\n",
"\n",
"The directory could in practice be used to store other data that should be accessible after the experiment is finished.\n",
"```python\n",
"# Use this module to get the TensorBoard logdir\n",
"from hops import tensorboard\n",
"tensorboard_logdir = tensorboard.logdir()\n",
"```\n",
"\n",
"### Using the `hdfs` module\n",
"The `hdfs` module provides a method to get the path in HopsFS where your data is stored, namely by calling `hdfs.project_path()`. The path resolves to the root path for your project, which is the view that you see when you click `Data Sets` in HopsWorks. To point where your actual data resides in the project you to append the full path from there to your Dataset. For example if you create a mnist folder in your Resources Dataset, the path to the mnist data would be `hdfs.project_path() + 'Resources/mnist'`\n",
"\n",
"```python\n",
"# Use this module to get the path to your project in HopsFS, then append the path to your Dataset in your project\n",
"from hops import hdfs\n",
"project_path = hdfs.project_path()\n",
"```\n",
"\n",
"```python\n",
"# Downloading the mnist dataset to the current working directory\n",
"from hops import hdfs\n",
"mnist_hdfs_path = hdfs.project_path() + \"Resources/mnist\"\n",
"local_mnist_path = hdfs.copy_to_local(mnist_hdfs_path)\n",
"```\n",
"\n",
"### Documentation\n",
"See the following links to learn more about running experiments in Hopsworks\n",
"\n",
"- Learn more about experiments\n",
" \n",
"- Building End-To-End pipelines\n",
" \n",
"- Give us a star, create an issue or a feature request on Hopsworks github\n",
"\n",
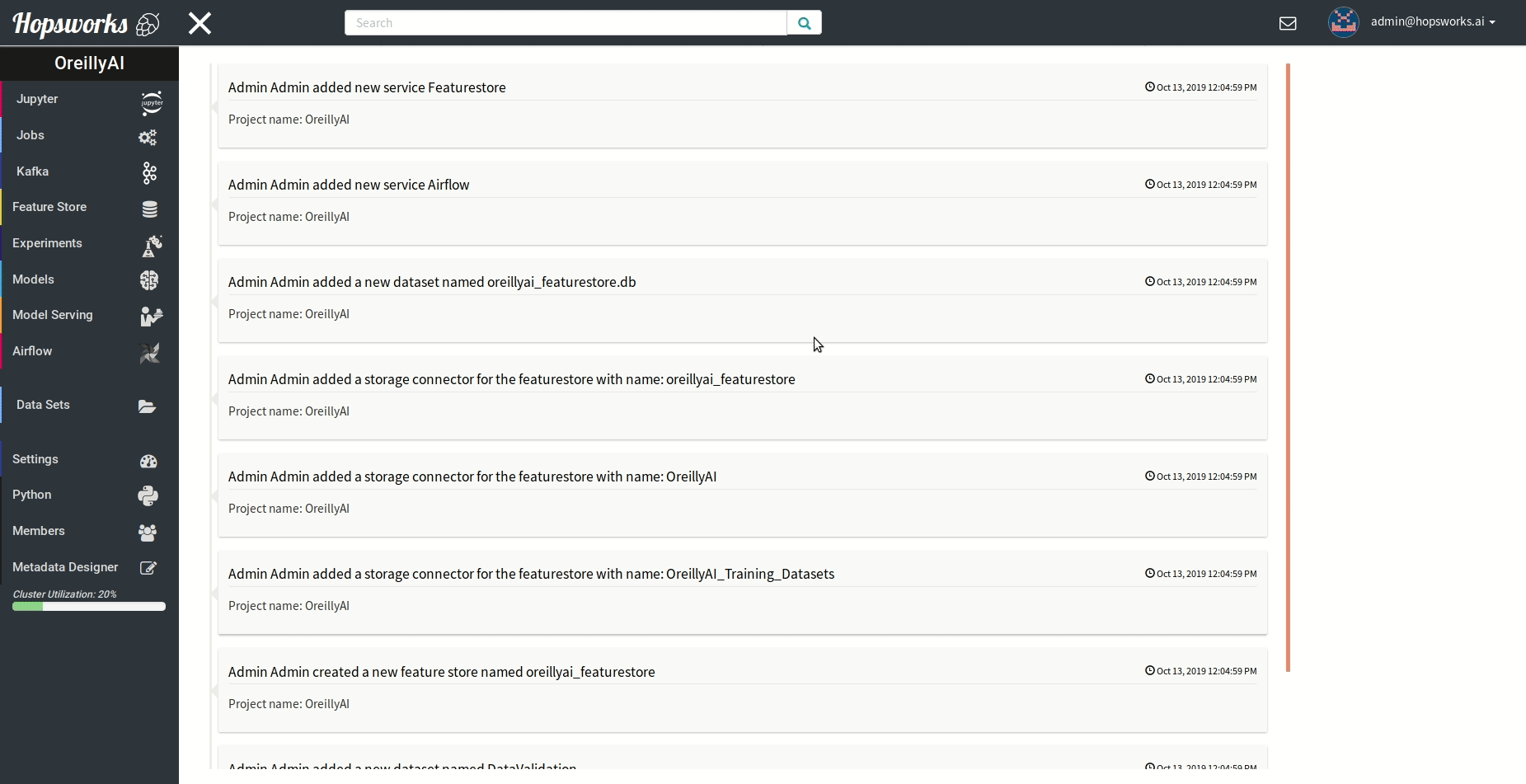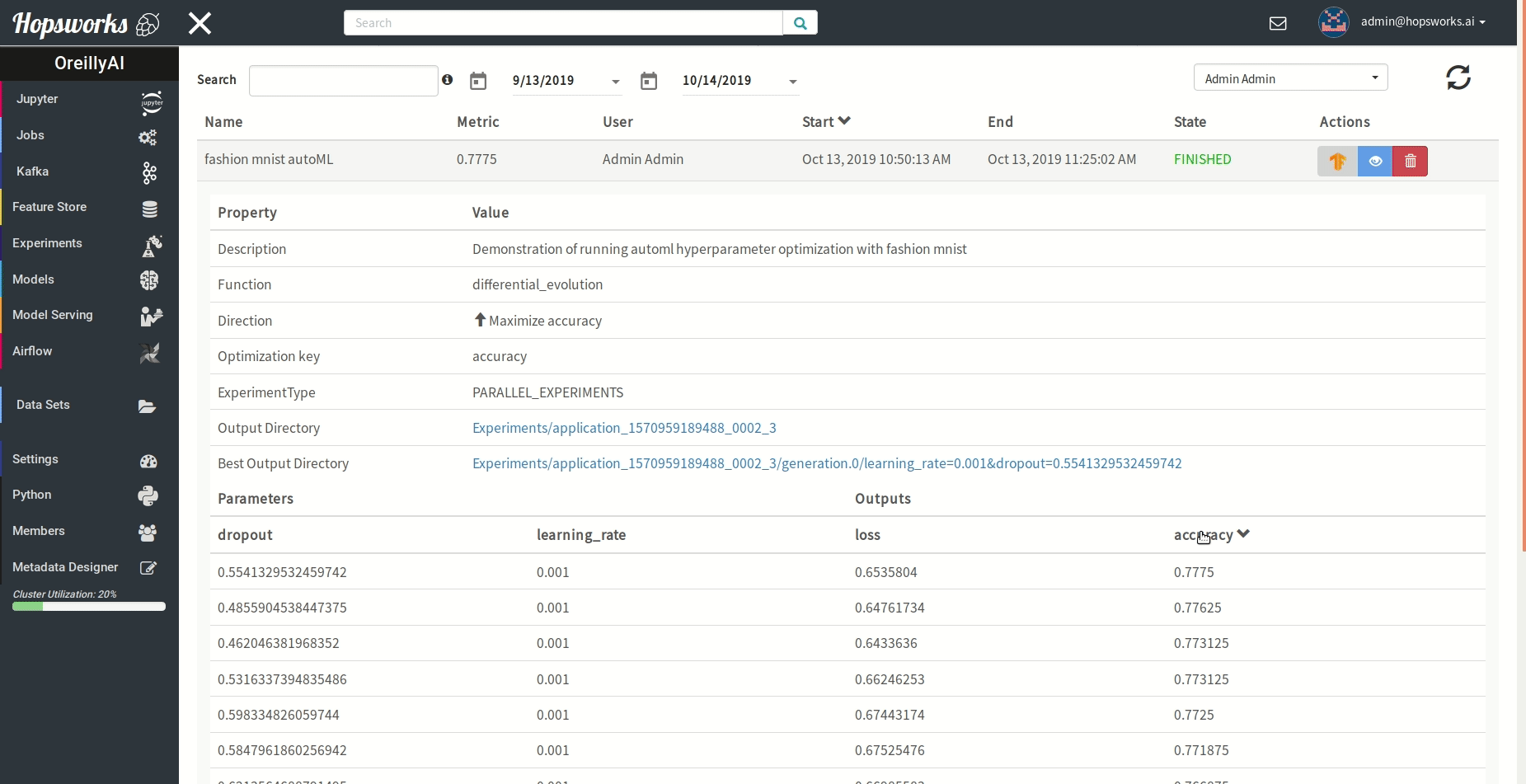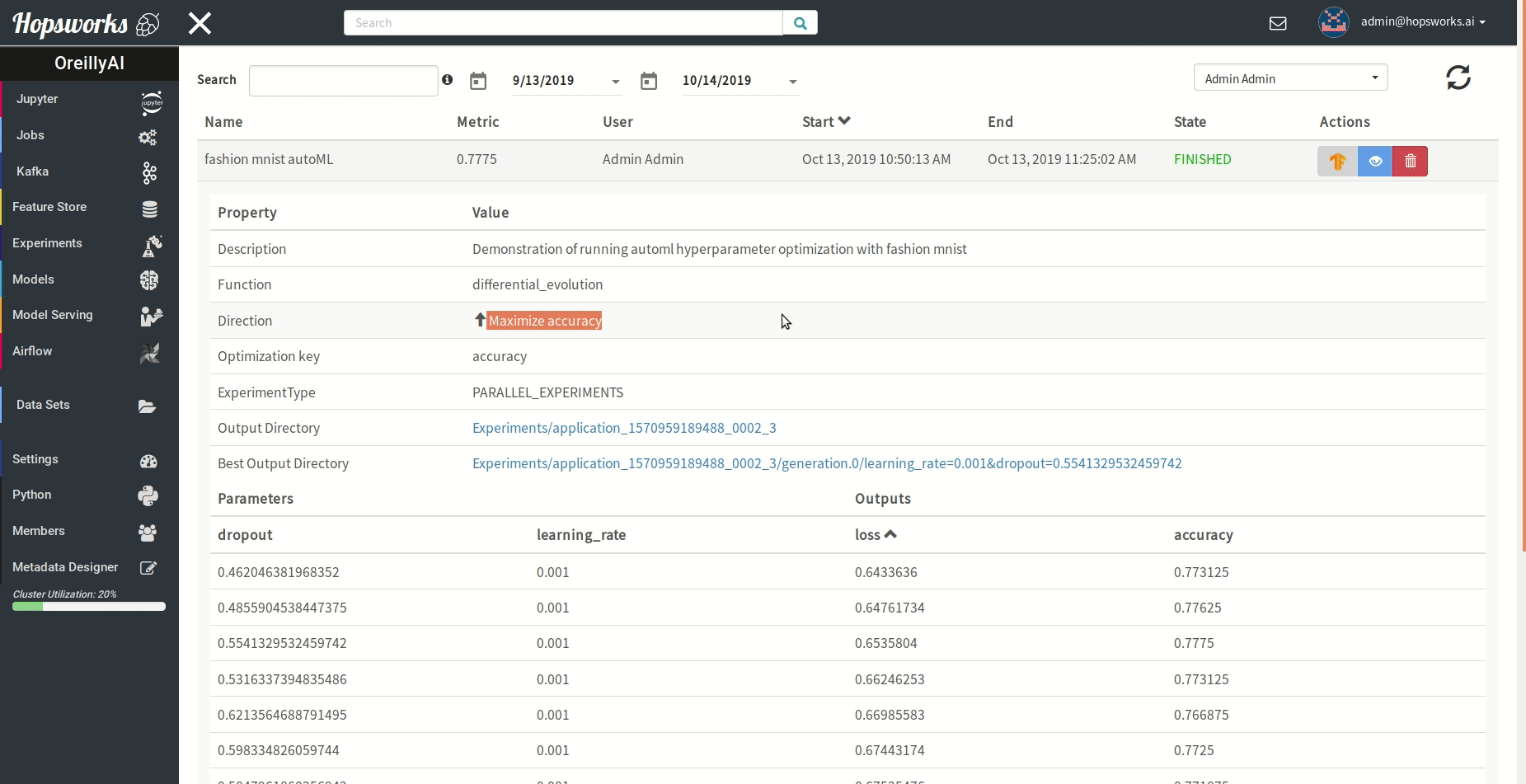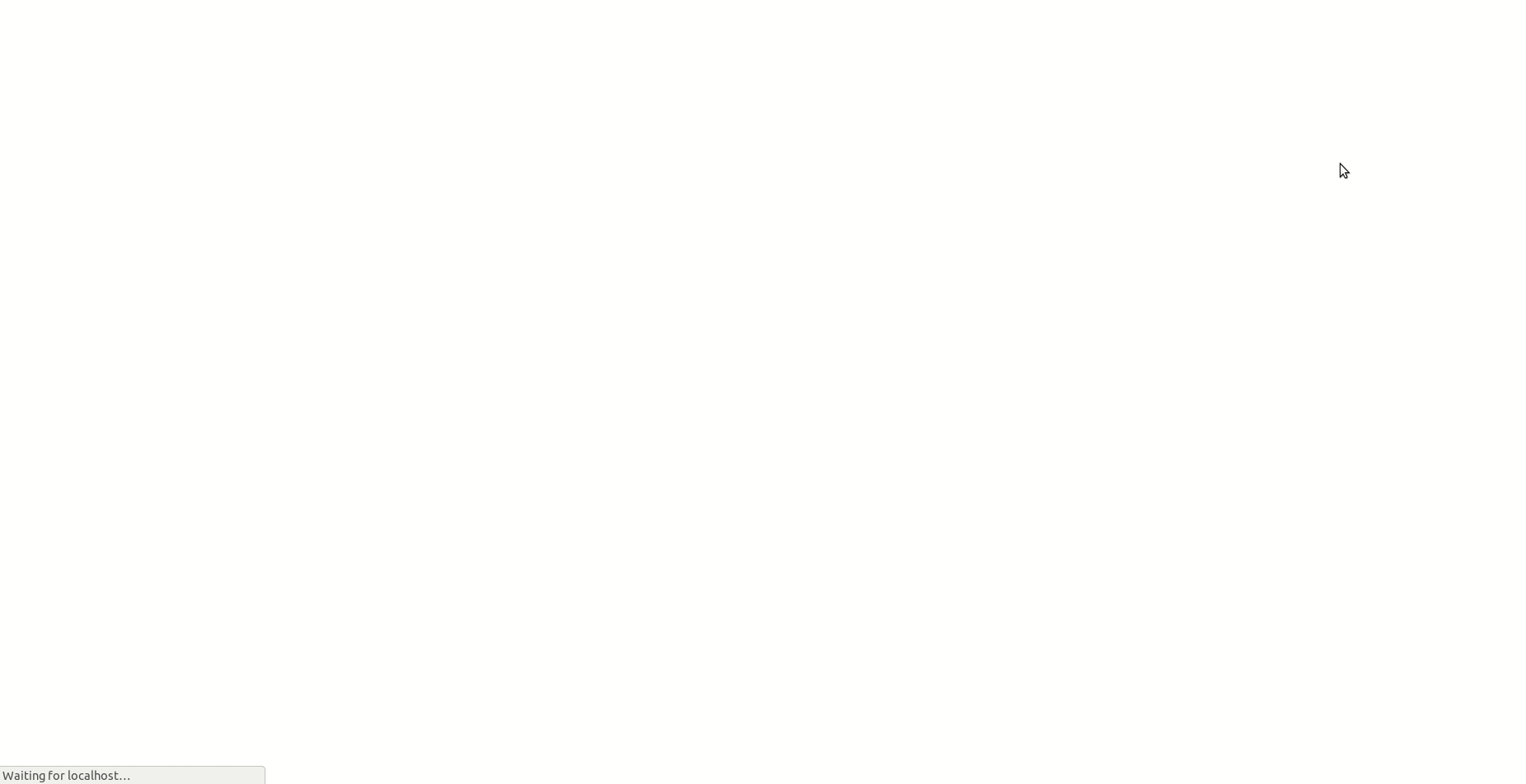
"### Managing experiments\n",
"Experiments service provides a unified view of all the experiments run using the `experiment` module.\n",
" \n",
"As demonstrated in the gif it provides general information about the experiment and the resulting metric. Experiments can be visualized meanwhile or after training in a TensorBoard.\n",
" \n",
" \n",
""
]
},
{
"cell_type": "code",
"execution_count": 1,
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Starting Spark application\n"
]
},
{
"data": {
"text/html": [
"