\n",
"\n",
"\n",
"\n",
"\n",
"## The `hops` python module\n",
"\n",
"`hops` is a helper library for Hops that facilitates development by hiding the complexity of running applications and iteracting with services.\n",
"\n",
"Have a feature request or encountered an issue? Please let us know on github.\n",
"\n",
"### Using the `experiment` module\n",
"\n",
"To be able to run your Machine Learning code in Hopsworks, the code for the whole program needs to be provided and put inside a wrapper function. Everything, from importing libraries to reading data and defining the model and running the program needs to be put inside a wrapper function.\n",
"\n",
"The `experiment` module provides an api to Python programs such as TensorFlow, Keras and PyTorch on a Hopsworks on any number of machines and GPUs.\n",
"\n",
"An Experiment could be a single Python program, which we refer to as an **Experiment**. \n",
"\n",
"Grid search or genetic hyperparameter optimization such as differential evolution which runs several Experiments in parallel, which we refer to as **Parallel Experiment**. \n",
"\n",
"ParameterServerStrategy, CollectiveAllReduceStrategy and MultiworkerMirroredStrategy making multi-machine/multi-gpu training as simple as invoking a function for orchestration. This mode is referred to as **Distributed Training**.\n",
"\n",
"### Using the `tensorboard` module\n",
"The `tensorboard` module allow us to get the log directory for summaries and checkpoints to be written to the TensorBoard we will see in a bit. The only function that we currently need to call is `tensorboard.logdir()`, which returns the path to the TensorBoard log directory. Furthermore, the content of this directory will be put in as a Dataset in your project's Experiments folder.\n",
"\n",
"The directory could in practice be used to store other data that should be accessible after the experiment is finished.\n",
"```python\n",
"# Use this module to get the TensorBoard logdir\n",
"from hops import tensorboard\n",
"tensorboard_logdir = tensorboard.logdir()\n",
"```\n",
"\n",
"### Using the `hdfs` module\n",
"The `hdfs` module provides a method to get the path in HopsFS where your data is stored, namely by calling `hdfs.project_path()`. The path resolves to the root path for your project, which is the view that you see when you click `Data Sets` in HopsWorks. To point where your actual data resides in the project you to append the full path from there to your Dataset. For example if you create a mnist folder in your Resources Dataset, the path to the mnist data would be `hdfs.project_path() + 'Resources/mnist'`\n",
"\n",
"```python\n",
"# Use this module to get the path to your project in HopsFS, then append the path to your Dataset in your project\n",
"from hops import hdfs\n",
"project_path = hdfs.project_path()\n",
"```\n",
"\n",
"```python\n",
"# Downloading the mnist dataset to the current working directory\n",
"from hops import hdfs\n",
"mnist_hdfs_path = hdfs.project_path() + \"Resources/mnist\"\n",
"local_mnist_path = hdfs.copy_to_local(mnist_hdfs_path)\n",
"```\n",
"\n",
"### Documentation\n",
"See the following links to learn more about running experiments in Hopsworks\n",
"\n",
"- Learn more about experiments\n",
" \n",
"- Building End-To-End pipelines\n",
" \n",
"- Give us a star, create an issue or a feature request on Hopsworks github\n",
"\n",
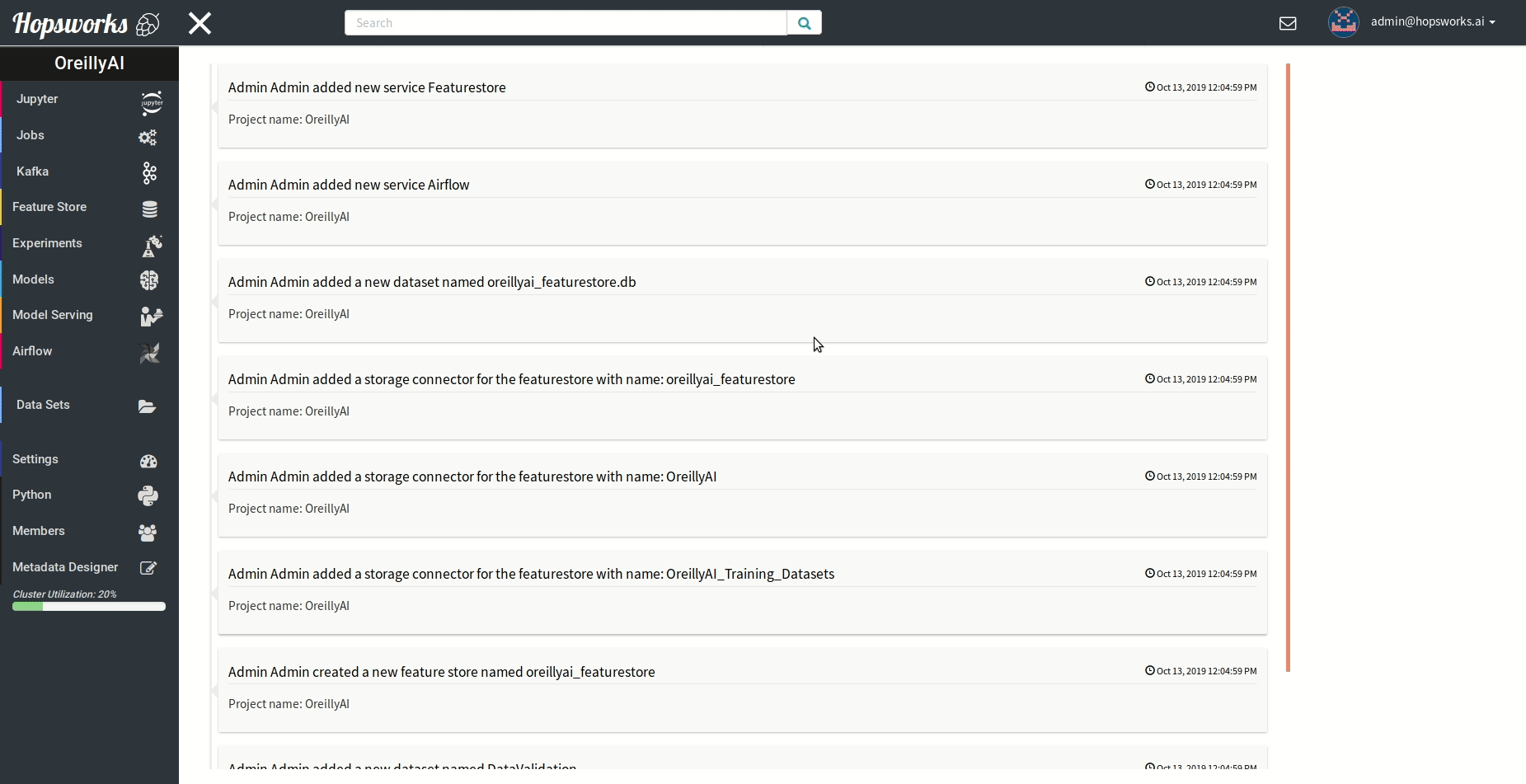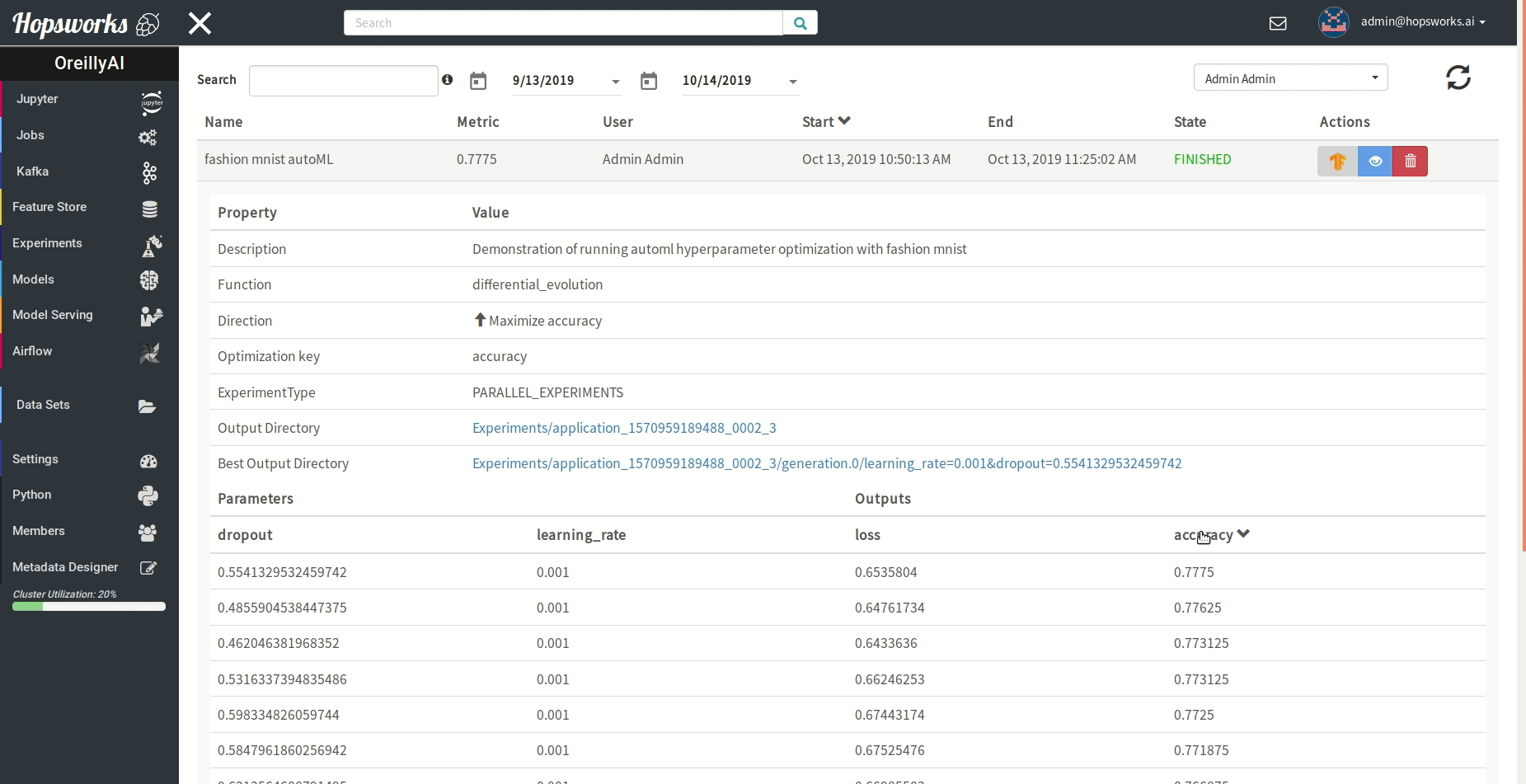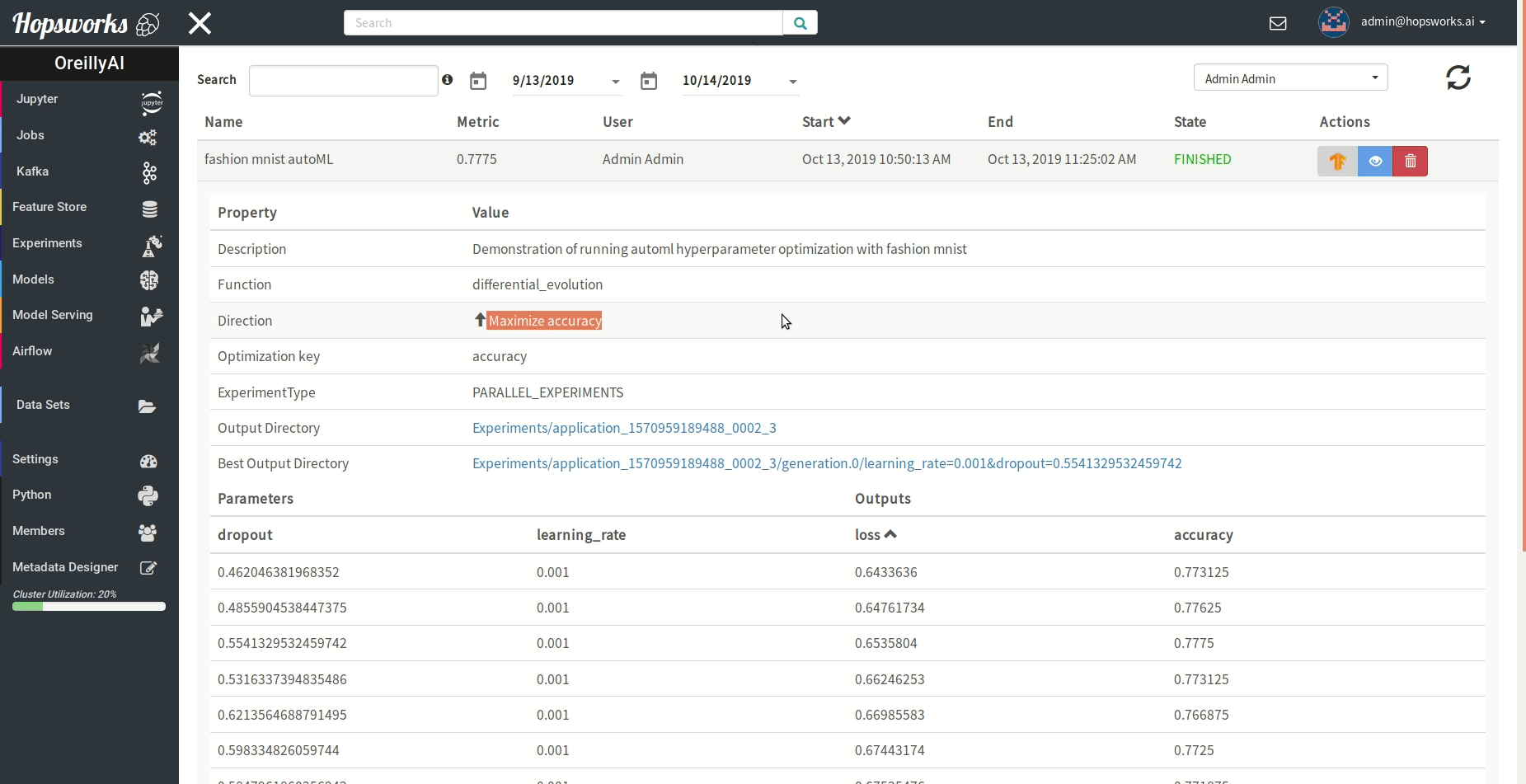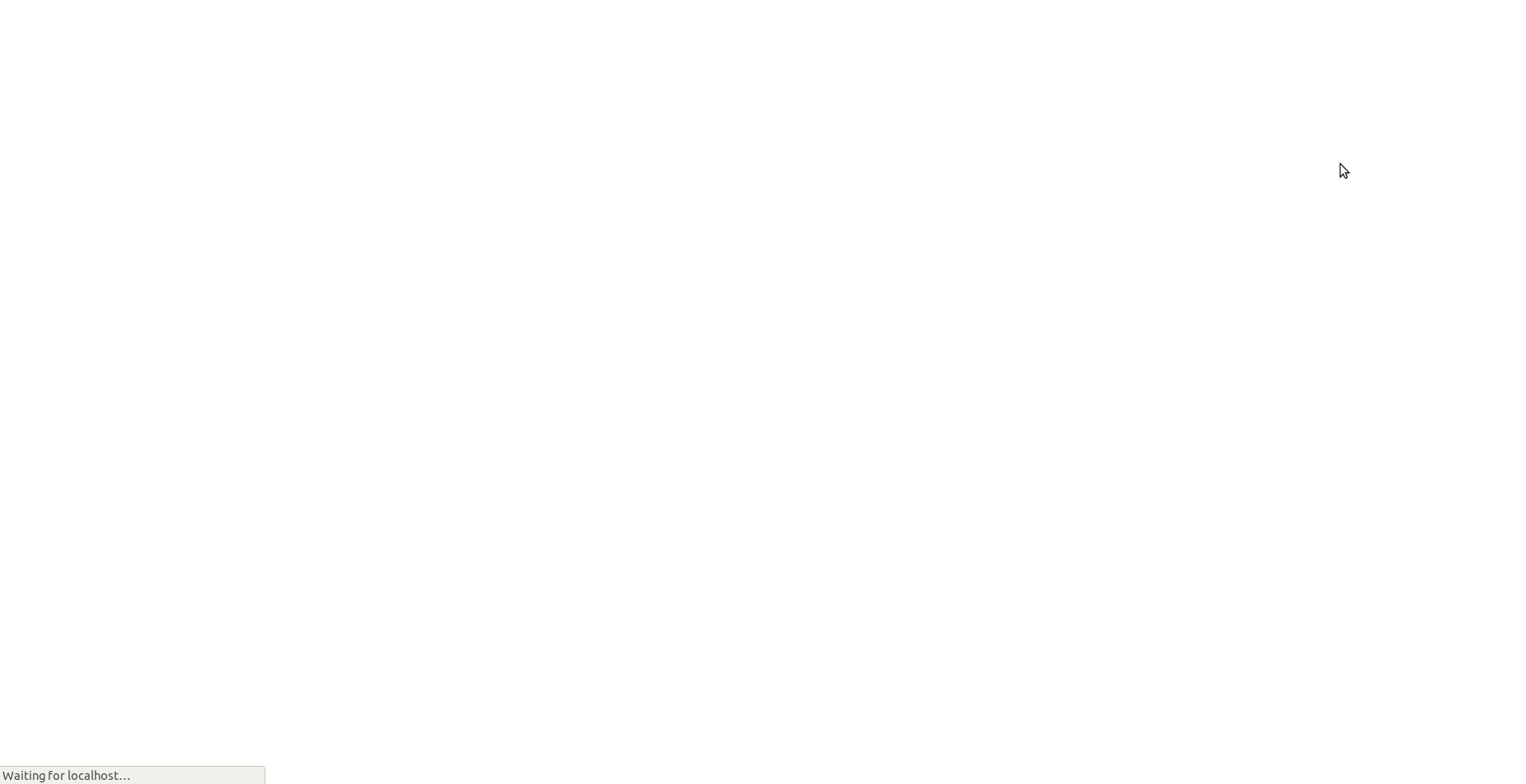
"### Managing experiments\n",
"Experiments service provides a unified view of all the experiments run using the `experiment` module.\n",
" \n",
"As demonstrated in the gif it provides general information about the experiment and the resulting metric. Experiments can be visualized meanwhile or after training in a TensorBoard.\n",
" \n",
" \n",
""
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"def keras_mnist():\n",
" from tensorflow.python import keras\n",
" import tensorflow as tf\n",
" from tensorflow.python.keras.datasets import mnist\n",
" from tensorflow.python.keras.models import Sequential\n",
" from tensorflow.python.keras.layers import Dense, Dropout, Flatten\n",
" from tensorflow.python.keras.layers import Conv2D, MaxPooling2D\n",
" from tensorflow.python.keras.callbacks import TensorBoard\n",
" from tensorflow.python.keras import backend as K\n",
"\n",
" import math\n",
" from hops import tensorboard\n",
"\n",
" batch_size = 8\n",
" num_classes = 10\n",
" epochs = 3\n",
" kernel = 4\n",
" pool = 4\n",
" dropout = 0.5\n",
"\n",
" # Input image dimensions\n",
" img_rows, img_cols = 28, 28\n",
"\n",
" # The data, shuffled and split between train and test sets\n",
" (x_train, y_train), (x_test, y_test) = mnist.load_data()\n",
"\n",
" if K.image_data_format() == 'channels_first':\n",
" x_train = x_train.reshape(x_train.shape[0], 1, img_rows, img_cols)\n",
" x_test = x_test.reshape(x_test.shape[0], 1, img_rows, img_cols)\n",
" input_shape = (1, img_rows, img_cols)\n",
" else:\n",
" x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)\n",
" x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)\n",
" input_shape = (img_rows, img_cols, 1)\n",
"\n",
" x_train = x_train.astype('float32')\n",
" x_test = x_test.astype('float32')\n",
" x_train /= 255\n",
" x_test /= 255\n",
" print('x_train shape:', x_train.shape)\n",
" print(x_train.shape[0], 'train samples')\n",
" print(x_test.shape[0], 'test samples')\n",
"\n",
" # Convert class vectors to binary class matrices\n",
" y_train = keras.utils.to_categorical(y_train, num_classes)\n",
" y_test = keras.utils.to_categorical(y_test, num_classes)\n",
"\n",
" model = Sequential()\n",
" model.add(Conv2D(32, kernel_size=(kernel, kernel),\n",
" activation='relu',\n",
" input_shape=input_shape))\n",
" model.add(Conv2D(64, (kernel, kernel), activation='relu'))\n",
" model.add(MaxPooling2D(pool_size=(pool, pool)))\n",
" model.add(Dropout(dropout))\n",
" model.add(Flatten())\n",
" model.add(Dense(128, activation='relu'))\n",
" model.add(Dropout(dropout))\n",
" model.add(Dense(num_classes, activation='softmax'))\n",
"\n",
" opt = keras.optimizers.Adadelta(1.0)\n",
"\n",
" model.compile(loss=keras.losses.categorical_crossentropy,\n",
" optimizer=opt,\n",
" metrics=['accuracy'])\n",
"\n",
" tb_callback = TensorBoard(log_dir=tensorboard.logdir(), histogram_freq=0,\n",
" write_graph=True, write_images=True)\n",
" callbacks = [tb_callback]\n",
" callbacks.append(keras.callbacks.ModelCheckpoint(tensorboard.logdir() + '/checkpoint-{epoch}.h5'))\n",
"\n",
" model.fit(x_train, y_train,\n",
" batch_size=batch_size,\n",
" callbacks=callbacks,\n",
" epochs=epochs,\n",
" verbose=1,\n",
" validation_data=(x_test, y_test))\n",
" score = model.evaluate(x_test, y_test, verbose=0)\n",
" return {'accuracy': score[1], 'loss': score[0]}"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": [
"from hops import experiment\n",
"from hops import hdfs\n",
"\n",
"experiment.launch(keras_mnist, name='keras mnist', local_logdir=True, metric_key='accuracy')"
]
},
{
"cell_type": "code",
"execution_count": null,
"metadata": {},
"outputs": [],
"source": []
}
],
"metadata": {
"kernelspec": {
"display_name": "PySpark",
"language": "",
"name": "pysparkkernel"
},
"language_info": {
"codemirror_mode": {
"name": "python",
"version": 2
},
"mimetype": "text/x-python",
"name": "pyspark",
"pygments_lexer": "ipython3"
}
},
"nbformat": 4,
"nbformat_minor": 4
}